The more we learn about the world around us, the more resources we have to make life better. From plasmids to CRISPR, we continue to take discoveries from the natural world and utilize them for further discoveries and treatments. Our “My Favorite Building Block” series takes the individual pieces of the castles we build in molecular biology and examines them in detail.
Before we begin looking at the pieces in depth, let’s take a moment to step back and set the stage by reviewing some basics in cloning and plasmids.
Plasmids
In order to introduce or modulate genes, it typically begins with a plasmid. These pieces of bacterial DNA were originally discovered in the 1960’s as a method of gene transfer between bacterial cells, but by the 1970’s, biologists were able to introduce genetic material to plasmids (Figure 1). These recombinant plasmids allowed scientists to introduce genes and manipulate gene expression in bacterial cells.
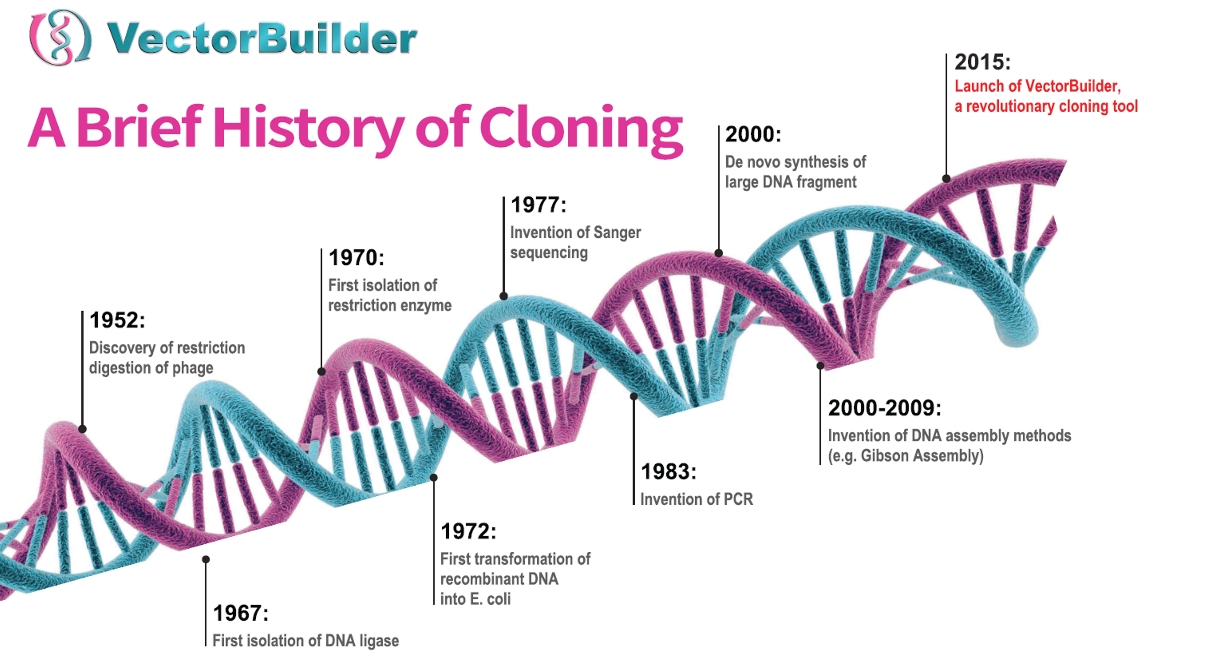
Figure 1. A brief history of major events in cloning
Making recombinant plasmids
The steps required to integrate DNA into plasmids were each independent discoveries that came together to transform the field of molecular biology. Studies in the 1960’s of bacterial cells that were able to chop up invading viral DNA led to the discovery and isolation of restriction endonucleases or restriction enzymes. These proteins bind to specific DNA sequences in any organism and introduce cuts. Once the two DNA fragments are together, they need to be joined, or ligated (Figure 2). This is facilitated by proteins, called ligases, which were discovered in the 1960’s. These breakthroughs all occurred within 10-15 years of the discovery of the structure of DNA in labs that would be unrecognizable to most scientists and students today.
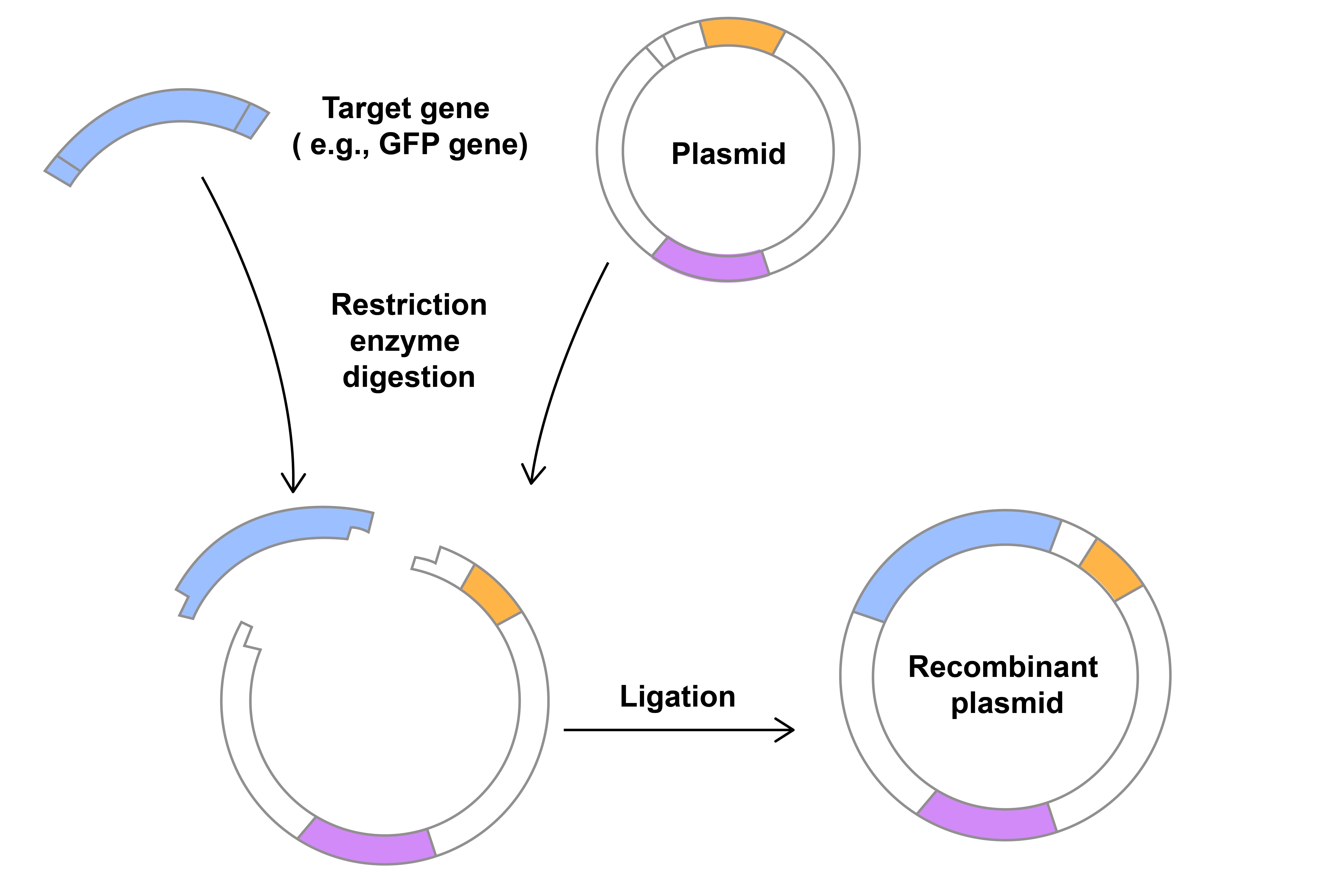
Figure 2. Digestion and ligation to insert a gene into a plasmid
After recombinant plasmids are created, they need to be introduced to a bacterial cell where the plasmids can be replicated as the host cells proliferate. Because of the lipid bilayer around cells, large molecules like plasmids cannot enter into cells unaided. Instead, a transformation technique is utilized, where competent bacterial cells are made temporarily permeable due to brief shock using either heat or electricity.
Making DNA
In modern molecular biology, we often need more than restriction enzymes to efficiently introduce genetic material into plasmids. Biologists are able to create millions of copies of DNA fragments and even introduce changes to those fragments through polymerase chain reaction (PCR). In PCR, short pieces of DNA oligos (primers) are designed that are complementary to both ends of your region of interest. These primers bind to their complementary sequence on DNA, and initiate replication by DNA polymerase. This process repeats over and over, with each new strand being a new template for the primers (Figure 3). The continued binding and extension produces millions to billions of copies of your sequence of interest in just a few hours. This PCR amplified fragment of DNA can then quickly be ligated into a plasmid to create endless possibilities for recombinant plasmids.
Figure 3. Steps in polymerase chain reaction (PCR)
A revolution in gene delivery
These molecular techniques allow us to isolate and clone sequences into plasmids, but in order to study their function, particularly in different organisms, this is only half the battle. While some bacterial strains can accept the plasmid through transformation, those bacteria often cannot infect other cells (e.g. plant or animal cells) and introduce the plasmids. Studying genes in other organisms typically requires further steps for introduction of genetic material.
In some cases, plasmids may be directly introduced into cells through transfection methods, those that do not utilize a virus. These include electroporation, where cells are shocked and become permeable to plasmid DNA, and lipofection, where plasmids are contained within a lipid bilayer which merges with the host cell. However, transfection is largely limited to in vitro or developmental models, and it generally cannot be used in differentiated and non-dividing cells.
Transduction, the use of viruses to introduce genetic material to cells, offers a wide range of options each with their own advantages and disadvantages depending on the specific viral system used. To generate the recombinant virus for transduction, typically one or more plasmids containing part of the viral genome as well as the gene(s) of interest are transfected into packaging cells, e.g. human embryonic kidney cells (HEK293). Following transfection, the cellular machinery uses the viral genome to create recombinant viruses that contain the gene(s) of interest, and the virus can then be isolated, purified and used to transduce target cells both in vitro and in vivo.
There are many options for types of viral vectors: lentivirus, adenovirus, AAV, retrovirus, and more. Each of these viruses have different characteristics, advantages, and disadvantages for gene delivery, so there are many factors to consider. Different viruses have widely varying capacity to carry and deliver your gene of interest, from about 4.7kb in AAV to 30kb in Vaccinia. Additionally, viruses interact with their host cells differently. Tropism, the breadth of target cells, varies from dividing to non-dividing cells, differentiated to stem cells, and primary vs passaged cells. Once in the cell, some viruses maintain the foreign genetic material as extrachromosomal, or episomal, DNA while others integrate into the host genome. Finally, viruses interact with the entire host organism to varying degrees: some risk damaging the host (pathogenicity) and some induce a host immune response (immunogenicity), both of which can obscure experimental results if not controlled for appropriately.
This series will take a deep dive into components that are critical to any and all of the processes we have described here: PCR, control of gene expression, experimental design, and more. We look forward to deconstructing the cloning castle with you.
Sources
Arber W. Host-controlled modification of bacteriophage. Annu Rev Microbiol. 1965;19:365-78. doi: 10.1146/annurev.mi.19.100165.002053. PMID: 5318444.
Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240-4. doi: 10.1073/pnas.70.11.3240. PMID: 4594039; PMCID: PMC427208.
Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557-80. doi: 10.1016/s0022-2836(83)80284-8. PMID: 6345791.
Hasan MM, Ragnarsson L, Cardoso FC, Lewis RJ. Transfection methods for high-throughput cellular assays of voltage-gated calcium and sodium channels involved in pain. PLoS One. 2021 Mar 5;16(3):e0243645. doi: 10.1371/journal.pone.0243645. PMID: 33667217; PMCID: PMC7935312.
Jackson DA, Symons RH, Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2904-9. doi: 10.1073/pnas.69.10.2904. PMID: 4342968; PMCID: PMC389671.
Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790-7. doi: 10.1126/science.186.4166.790. PMID: 4377758.
Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51 Pt 1:263-73. doi: 10.1101/sqb.1986.051.01.032. PMID: 3472723.
Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982;1(7):841-5. doi: 10.1002/j.1460-2075.1982.tb01257.x. PMID: 6329708; PMCID: PMC553119.
Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737-8. doi: 10.1038/171737a0. PMID: 13054692.



